
Biologist Attila Reményi, scientific advisor at the Research Centre for Natural Sciences of the Hungarian Academy of Sciences (RCNS-HAS)
Your project proposal submitted to the Frontline Call reflects that you see your work in the coming years as part of a long-term vision.
The groups of proteins, i.e. the enzymes called MAP kinase that I am working with play a central role in cell division (for example in cancer diseases) and in cell death (in chronic inflammatory processes).
I started this research during my postdoctoral studies in 2002. At that time, I took yeast as a model organism to see how cells process ambient physical and chemical stimuli and how they give biological responses that are crucial to their survival. To understand the laws among proteins, I drew up an atomic-level map of the proteins turned off and on (activated). In a following project we made atomic-level molecular maps that contributed to explaining the behaviour of human cells and, owing to that, I could establish my laboratory under the Momentum Programme.
The idea of using molecules with which we can intervene in the complex processes transducing external signals arouse already at that time as we had acquired very detailed information on proteins needed for signal transduction. If we could design an active ingredient capable of breaking the connection between two proteins, that would give us a chance of pharmaceutical drugs to be developed later on, we supposed. Leaving the atomic level behind and jumping to a much higher level is at the heart of the project submitted to the Frontline Call. All the micro-level information acquired about the activities on atomic level will be used for designing experiments whereby we can globally interpret signal transduction and the functioning of interrelated signal pathways. To this end we carry out targeted small interventions.
And eventually from the atoms you will get to the system as a whole. This knowledge may even serve as the basis of medical treatments.
Our primary duty is providing new directions: we need to point out how more efficient active ingredients can be produced through understanding a phenomenon. Development of pharmaceutical drugs will then be the business of pharmaceutical companies. We should map which proteins interact with each other and which ones “turn on” others. The heart of our Frontline proposal is the idea of a “designer cell”: we created cells in which, if I wish, I can turn on either the enzyme responsible for cell division only or the one responsible for cell death. We are trying to scan the signal’s pathway right inside the cell.
How can you do that?
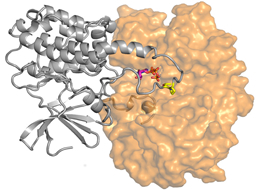
Molecular image of protein interaction in signal transduction.
Having such atomic-level information allows a deeper understanding of cell biological processes. The image depicts the moment when a MAP kinase called ERK (orange) and an executing protein triggering cell growth (grey) meet.
I have molecular images, for example, of the protein that activates the MAP kinase (the enzyme involved in cell division) and I can check what processes they govern in the cell, for example during cell division or cell death. No such refined analyses have been made so far because the individual relationships among proteins were not adequately looked into. We would like to intervene in the processes knowingly and that would also be the basis for tailored pharmacotherapy. That is the reason why I am getting more and more involved in projects aiming to develop targeted therapies, such as Balázs Győrffy’s project under the National Competitiveness and Excellence Programme (this project has a world-class unique approach using a computer algorithm to analyse the “initial” gene defects of solid brain tumour in children). It is important to test the active ingredients under controlled circumstances.
This is largely supported by an infrastructural background, which also rests on your commitment for cooperation.
I happened to represent Hungary in a big chemical biology consortium called EU-Openscreen (EU-Openscreen is a consortium set up by biological and pharmaceutical chemistry research centres of 17 member states which Hungary joined in 2013. The consortium undertook to establish – starting from 2016 – a library of compounds carefully considered from the aspect of molecular pharmacology screening). In addition to ELI, the European Research Infrastructure Consortium (ERIC) whose task is to facilitate the establishment of infrastructures included, among others, EU-Openscreen in its programme. It doesn’t mean though that a single large infrastructure is being established but several smaller ones, in various countries. These will be high-capacity automated systems testing large numbers of molecules serving the purposes of active ingredient development.
At the sessions organised for building up the professional background of the chemical biology consortium, I found that in infrastructure establishment “the roles are already taken” but there are still niches where we can contribute to the consortium’s work on national level. I am working towards this through a specific project financed under the Competitive Central Hungary Operational Programme (CCHOP/VEKOP). We would like to develop a system that can measure the strength of protein associations and the speed of processes, and also gain insight into these inside the cells. This approach is really rare. Our results could be used by researchers for optimizing active ingredients in the future. In this project we have to put the infrastructure into operation but I also agreed to establish a library of molecules (collecting and cataloguing compounds) which will be available to the researcher community. With this acedemic library we can join the big library of molecules to be established by EU-Openscreen.
How many people do you have in your team for all this work?
Three senior researchers, a postdoctoral researcher, five PhD students and a laboratory technician, so we are eleven in the team. Thus the research process is followed also by “science management”: today I have to think in units and cooperation beyond the level of my own group. It is a challenge and responsibility but a new opportunity at the same time.






